Author: Sydney Wyatt
Editor: Keith Fraga
Human genetic modification strikes fear into many scientists and non-scientists alike. The recent claim of human genetic editing experiments suggested that a researcher in China, Dr. He Jiankui, edited human embryos to be resistant to HIV, and that some embryos were successfully carried to term (1). While the twin girls, and a potential third baby on the way from another couple, seem to be healthy, this may not be the first time Chinese researchers have genetically modified humans (2).
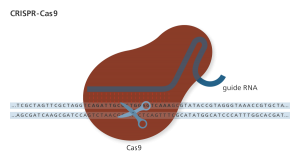
An RNA guide targets the Cas9 nuclease through complementary binding. The variable guide length allows for relatively easy design with high specificity to the target site. Genome Research Limited
Since the advent of CRISPR*, a specific genetic editing tool derived from a native bacteria defense system, genetic modification is the new hot ticket for research and media coverage (3). The technology is so accessible, citizen-scientists popularly known as biohackers have attempted to modify themselves with DIY CRISPR kits (4). While self-experimentation is discouraged, it begs the question: should scientists genetically modify a human using CRISPR?

Process of using recombinant DNA to engineer E. coli to produce human insulin for mass production. Genome Research Limited
Genetic editing is not a new technology. Recombinant DNA** has existed since the 1970s and is used extensively in research and biotechnology companies to create genetically modified organisms (5). For example, Genentech and Eli Lilly & Co used recombinant DNA to genetically modify bacteria in order to mass produce human insulin used to manage diabetes (approved by the FDA in 1982). Genetically modifying bacteria dramatically increased yield and purity of insulin over animal-sourced insulin, improving the access to this vital therapeutic for insulin patients.
At the time of its discovery, there was concern within the science community as well as within the general public over gene manipulation in humans using recombinant DNA. In February 1975, biologists, lawyers, and journalists gathered for the second Asilomar Conference on Recombinant DNA to draft regulations on experiments using recombinant DNA technology (6). Just prior to the conference, a moratorium on research projects using recombinant DNA had been voluntarily put in place and universally observed — a remarkable example of scientists’ ability to self-regulate (7). The recommendations that emerged from the conference addressed “how the scientific work could be undertaken with minimal risks to workers in laboratories, to the public at large, and to the animal and plant species sharing our ecosystems.”
Ultimately, the safety precautions were laid out in guidelines issued by the NIH in July 1976 but never became law despite legislators’ suggestions. Researchers instead continued self-regulation with the added guidance of the NIH Recombinant DNA Advisory Committee. It seemed like excessive red tape to deal with in order to perform any experiments using this technology, but the regulations were necessary to ensure public trust in scientific endeavors (7). In the decades since, these regulations have been refined to address new concerns in the field while maintaining forward momentum.
The far-reaching effects of the 1975 Asilomar Conference remains unmatched today. The new tech on the block is CRISPR, which is a far more nuanced and powerful genetic editing tool than recombinant DNA.
CRISPR/Cas9, commonly referred to as CRISPR in the media, is an engineered nuclease. And it’s not the first. Meganucleases (c. 1980s), zinc finger nucleases (ZFN; c. 1990s), and transcription activator-like effector nucleases (TALEN; c. 2010) make up the larger family of engineered nucleases. ZFNs, TALENs, and CRISPR (c. 2013) have been used successfully for genetic editing (8).
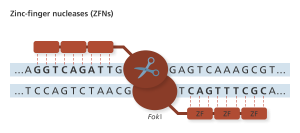
Zinc fingers (ZF) recognize codons, or 3-base codes in the DNA, to guide the FokI nuclease to the cut site. Genome Research Limited
ZFNs were the first truly programmable genetic editing tool and was applied in humans to disrupt the CCR5 co-receptor that HIV uses to enter cells. Preclinical trials using ex vivo somatic cell gene editing successfully demonstrated that using ZFN gene therapy for HIV treatment was feasible and safe and thus merited clinical trials with the intent to take this treatment to market (9). Phase I and Phase II clinical trials are being conducted in the United States regarding the treatment of HIV patients; clinical trials are also underway for several other diseases (10). Importantly, these trials edit only somatic cells of the patient, not the germline cells (eggs or sperm), so the changes made will not be inherited. I spoke with gene therapy researcher Dr. David Segal of University of California, Davis, about these clinical trials.
“The things that go into clinical trial, by government regulation, have to pass certain thresholds for safety and efficacy…We strive to say anything that goes into a person is as safe as we know it to be. So everyone should know [that] about clinical trials. They go to great lengths to demonstrate safety above all else – even before efficacy – when you go into clinical trial.” – Dr. David Segal
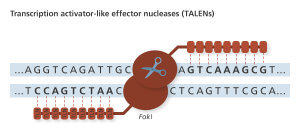
TLN sites recognize single bases, allowing for more freedom in choosing targets for editing with the FokI nuclease. Genome Research Limited
TALENs were next on the scene, but were quickly outshined by CRISPR. Thus, human genetic editing using TALENs has not made it to clinical trials in the United States.
This brings us full circle to CRISPR’d humans. It is interesting that He Jiankui, the man behind the CRISPR babies, also chose to target CCR5. Since the original MIT Technology Review article was published, a January 21 report from Xinhua, China’s state media agency, has confirmed the birth of genetically edited twin girls and the pregnancy of another couple, suggesting a third genetically edited could be born (11).
Was this ethical? According to the report, no. Segal elaborated on the ethical measures we have regarding genetic editing in humans and that He bypassed these ethical measures for his germline editing experiments:
“[W]hen science starts to push up against ethical boundaries, we have institutions in place to try to respect, society’s concerns about the research that are being done…it’s absolutely essential that all the work go through this committee…anything you do that involves people, even if it’s a psychological questionnaire that at the worst case could cost someone some mental harm, all the way up to working with human embryos…has to be approved by an institutional review board. And again, they want to maintain transparency…They are the oversight at this institution. They have different people on the committee. There’s kinds of protections that they look for. All the people doing the research need to have training in ethics of human research that involves the unethical behaviors that had been done in the past and how we need to avoid that. If we don’t train the investigators, if we don’t go through the IRB, if we don’t follow this procedures, these institutions that have been set up to maintain transparency and trust and the scientific endeavor in the eyes of society, the investigator could lose his job. The NIH can stop funding the entire university. I mean, there are big consequences [in] trying to circumvent these structures that have been put in place to maintain society’s confidence in the scientific ventures. And I would say that was the most egregious thing that scientist in China did.”
He Jiankui blatantly broke these agreements, resulting in the ethical uproar over his experiments. Segal also discussed the long-term consequences of germline gene editing and gene therapy in our interview, which can be read in full here.
Interestingly, a group of scientists, including a few of the original discoverers of CRISPR and recombinant DNA, issued a statement published in Science in 2015 requesting a moratorium on human genetic modification (12). Echos of the recommendations from the Asilomar Conference are present in the recommendations laid out in the statement. According to Segal, some of these recommendations have come to pass – CRISPR research transparency is highly encouraged through the establishment of forums like the International Summit on Human Genome Editing where He unveiled his work.
The recommendations: 1) Discourage, even in countries that might be permissive, any research aimed at heritable genetic modification; 2) Establish forums for information and education on the risks and rewards of using CRISPR to treat or cure human disease, and the accompanying ethical, social, and legal concerns; 3) Support research transparency to help determine whether or which clinical applications are permissible; 4) Gather experts in genetics, law, and bioethics as well as members of the scientific community and public at large to consider the issues at hand.
A publication from 2017 discussed similar concerns about whether we are prepared for CRISPR clinical trials (13). The publication mentions a Phase I clinical trial was already in progress in China with the intention of treating stage IV metastatic non-small cell lung cancer, but that a similar trial was still prospective in the United States. The authors offered an extensive and critical review of the preclinical data that was presented to the NIH Recombinant DNA Advisory Committee (sound familiar? It’s the same committee that was established around the time of the Asilomar Conference).
In short, they determined we are not ready. This conclusion was informed by an existing framework for assessing the jump from preclinical to clinical trials (14-17).
Yet on February 1, NPR published an article on human genetic editing that is happening in the United States (18). Dieter Egli of Columbia University claims that he is conducting his experiments genuinely for research, in contrast to He’s supposed goal of genetically protecting the babies from HIV. Currently, Egli is focused on correcting one of the underlying genetic defects that result in inherited blindness and only allows the modified embryos to develop for one day, though he hopes to allow further development if these initial experiments are successful.
Federal funds are banned from being used for this kind of research – germline editing – in the United States, but there is no such stipulation on private funding (i.e. self-funded, as implied in the case of He’s work). If you recall, DIY CRISPR kits are readily available online, so if one has the means, then there is nothing preventing DIY designer babies beyond one’s ethics. There is still considerable controversy: should the moratorium be instilled? Should it be applied to basic research, such as Egli’s project? Can adequate regulations be established to prevent the need for a moratorium?
According to Segal, “we’re in the very early days of trying to use this as a therapy. Most of the work doesn’t involve any humans, but some things are progressing to a point where it can be used in humans, and in clinical trial.”
Only time will tell. In the meantime, don’t fear the CRISPR. It won’t be coming to a human near you anytime soon.
Thank you Dr. Segal for taking the time to provide his expert opinion on this topic.
*CRISPR: a genetic engineering tool using a short, repetitive DNA sequence and associated editing protein Cas9 to specifically edit target DNA sequence.
**Recombinant DNA: DNA made by artificially combining DNA fragments from different organisms.
For more history on genetics, check out The Gene by Siddhartha Mukherjee.
Bibliography:
- Regalado, A. (2018, November 26). EXCLUSIVE: Chinese scientists are creating CRISPR babies.
- Foley, K. E. (2018, January 26). Chinese scientists used Crispr gene editing on 86 human patients.
- Doudna, J. (2015, January). Genome Engineering with CRISPR-Cas9: Birth of a Breakthrough Technology.
- Lee, S. M. (2019, January 17). This Biohacker Is Trying To Edit His Own DNA And Wants You To Join Him.
- Herbert W. Boyer and Stanley N. Cohen. (2017, December 11).
- Berg, P., Baltimore, D., Brenner, S., Roblin, R. O., III, & Singer, M. F. (1975). Summary Statement of the Asilomar Conference on Recombinant DNA Molecules. PNAS, 72(6), 1981-1984. doi:10.1073/pnas.72.6.1981
- The Paul Berg Papers: Recombinant DNA Technologies and Researchers’ Responsibilities, 1973-1980. (n.d.).
- Chandrasegaran, S., & Carroll, D. (2016). Origins of Programmable Nucleases for Genome Engineering. Journal of Molecular Biology, 428(5), 963-989. doi:10.1016/j.jmb.2015.10.014
- DiGiusto, D. L., Cannon, P. M., Holmes, M. C., Li, L., Rao, A., Wang, J., . . . Zaia, J. A. (2016). Preclinical development and qualification of ZFN-mediated CCR5 disruption in human hematopoietic stem/progenitor cells. Molecular Therapy — Methods & Clinical Development, 3. doi:10.1038/mtm.2016.67
- Search of: Zfn | United States – List Results. (n.d.).
- Cross, R. (2019, January 21). Rogue CRISPR scientist will be punished. C&EN, 97(3), 1-56.
- David, B., Berg, P., Botchan, M., Carroll, D., Charo, R. A., Church, G., . . . Yamamoto, K. R. (2015). A prudent path forward for genomic engineering and germline gene modification. Science,348(6230), 36-38. doi:10.1126/science.aab1028
- Baylis, F., & McLeod, M. (2017). First-in-human Phase 1 CRISPR Gene Editing Cancer Trials: Are We Ready? Current Gene Therapy, 17, 309-319. doi:10.2174/1566523217666171121165935
- Kimmelman J. (2009). Gene transfer and the ethics of first-in-human research: lost in translation. Cambridge University Press.
- Henderson V.C., Kimmelman J., Fergusson D., Grimshaw J.M., & Hackam D.G. (2013). Threats to validity in the design and conduct of preclinical efficacy studies: A systematic review of guidelines for in vivo animal experiments. PLoS Med, 10(7):e1001489.
- Kimmelman J., & Henderson V. (2015). Assessing risk/benefit for trials using preclinical evidence: a proposal. J. Med. Ethics, 42(1), 50.
- Kimmelman J., & London A.J. (2011). Predicting harms and benefits in translational trials: ethics, evidence, and uncertainty. PLoS Med, 8(3):e1001010.
- Stein, R. (2019, February 01). New U.S. Experiments Aim To Create Gene-Edited Human Embryos. Retrieved from https://www.npr.org/sections/health-shots/2019/02/01/689623550/new-u-s-experiments-aim-to-create-gene-edited-human-embryos