Author: Hongyan Hao
Editors: Anna Feitzinger | Emily Cartwright, Keith Fraga, Jessica Huang, Sharon Lee, Yulong Liu, Linda Ma
Long distance cargo transport in neurons is facilitated by microtubule motors kinesin and dynein. In vivo, microtubules are crowded environments covered by different kinds of microtubule associated proteins (MAPs). How do motors react when they encounter MAPs? A recent work entitled ‘competition between microtubule-associated proteins directs motor transport’ published in Nature Communications by Ph.D. student Brigette Monroy in the Biochemistry, Molecular, Cellular, Developmental Biology (BMCDB) program explored how different MAPs directs motor movement on the microtubules.
My ability to write this article depends on my nervous system formed by numerous cells called neurons. Neurons communicate with each other through a long cable-like projection termed an axon, and branched cellular extensions called dendrites. The axons of our motor neurons can extend all the way from the spinal cord to our toes, which can be 1 meter in length. Thus, neurons are faced with the significant challenge of transporting proteins, vesicles and organelles over long distances to maintain cell polarity and the connection between neurons.
Hollow tubes called microtubules (MTs) are the highways for effective intracellular transport from the axon or dendrite to the cell body. Motor proteins kinesin and dynein are the trucks traveling along the MTs. Polarized with two distinct ends, i.e. a plus end and a minus end, MTs are one-way-only tracks for both motors. Kinesin-1 is plus end directed while dynein moves towards the minus end of MTs. Thus, the organization of the MT is essential for directing cargos to their correct destination.
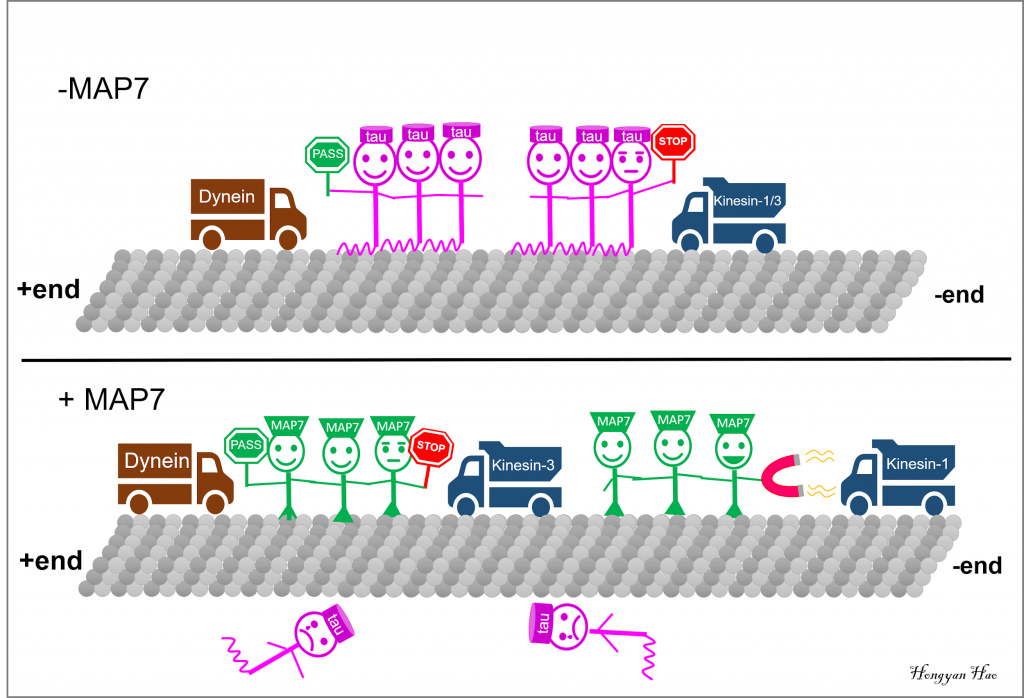 Microtubule (MT) plus-end motor kinesin-1 and kinesin-3 are inhibited by tau patches, while tau has little effect on the MT minus-end motor dynein. MAP7 outcompetes tau and specifically enhances kinesin-1 movement. Similar to tau, MAP7 inhibits kinesin-3 but allows for the dynein movement.
Microtubule (MT) plus-end motor kinesin-1 and kinesin-3 are inhibited by tau patches, while tau has little effect on the MT minus-end motor dynein. MAP7 outcompetes tau and specifically enhances kinesin-1 movement. Similar to tau, MAP7 inhibits kinesin-3 but allows for the dynein movement.
In axons, MTs are highly organized into bundles with plus ends directed out towards the cell periphery, and minus ends oriented inward towards the cell center. In contrast, MT orientation is mixed in dendrites (1). In vivo, MT organization and dynamics are highly regulated by microtubule associated proteins (MAPs). For example, the MAP tau binds to the MT lattice and stabilizes axon MTs. When detached from the MT, tau proteins form aggregates, which is one of the hallmarks of Alzheimer’s disease (2). Another MAP, MAP7, also known as ensconsin or EMAP-115, binds to MTs at axon branch sites to positively regulate branch formation (3). As the MTs are decorated by different kinds of MAPs in axons and dendrites, how do the motors dynein and kinesin behave when they hit MAPs?
MAPs are not just rocks on the road that block motors. In fact, the two motors will respond differently depending on which MAP they encounter. For instance, previous studies have shown that kinesin-1 motors will stop and fall off when they encounter tau, while tau has little effect on dynein (4, 5). In contrast, MAP7 enhances kinesin-1 movement (6, 7). How do these different motors and MAPs coordinate with each other to achieve effective cargo transport in axons and dendrites?
A recent publication by BMCDB student Brigette Monroy and her colleagues from Kassandra Ori-McKenny’s lab at UC Davis explores how motor movement might be regulated by different MAPs coating the MTs (8). Consistent with previous reports, Monroy et al. found that tau is restricted to axons, but MAP7 was localized to both axons and dendrites in the Drosophila peripheral nervous system and tissue cultured mouse neurons. Using Total Internal Reflection Fluorescence (TIRF) microscopy, which allows for visualization of molecules on a single stabilized MT in vitro, the authors found that MAP7 strongly excluded tau from the MTs. Although bound to MTs more rapidly than MAP7, tau is displaced by MAP7 over time. This is most likely due to MAP7’s higher MT-binding affinity and longer dwell time on the MT lattice since purified MAP7 and tau do not bind to each other (8).
It is striking that MAP7 kicks tau off the MTs, but what are the physiological functions of MAP7/tau competition? Overexpression of MAP7 in Drosophila larval dendritic arborization (DA) neurons caused increased branch number while tau overexpression led to a decreased branch number. Overexpression of both MAP7 and tau led to more branches in DA neurons, consistent with the observation that MAP7 outcompetes tau (8).
One possible mechanism is that MAP7 and tau affect DA neuron branching by regulating the intracellular transport of the Golgi outposts. Golgi outposts are important for branch formation by nucleating MTs at branch sites (3, 9). The increased branching observed with MAP7 overexpression may be due to increased kinesin-1 mediated cargo transportation since Golgi were found to be enriched in the plus ends of MTs in DA neurons.
In vivo studies have revealed positive regulation of kinesin-1 by MAP7, but the mechanism has been unclear (7). Using in vitro single molecule motility assays, where the movement of a single fluorescently labeled motor protein on the MT is observed under TIRF microscopy, Monroy et al. found that both human and Drosophila MAP7 directly affect kinesin-1 by enhancing the motor landing on the MT. This may be due to 34 conserved amino acids in MAP7 which may facilitate kinesin-1 recruitment through a weak, ionic interaction with kinesin-1. Kinesin-1 motors stop and fall off the MTs when they encounter patches of tau. Would the presence of MAP7 overcome tau’s inhibition on kinesin-1? The answer is yes. MAP7 can not only replace tau on the MT and recruit kinesin-1 onto the MT, but also restores the ATPase activity of kinesin-1 inhibited by tau.
Tau has little effect on dynein motors. What about MAP7? Interestingly, just like tau, MAP7 barely affects the mobility of the dynein active complex in vitro. So, does MAP7 only affect plus end MT motors? Kinesin-3 family motors are also plus end directional. In contrast to kinesin-1, MAP7 inhibits kinesin-3’s landing on the MT as well as its mobility. However, tau inhibits kinesin-3 in a similar way as kinesin-1.
Cells have very complex environments and it is challenging for in vivo studies to provide clear and detailed mechanisms of how proteins work with one another. On the other hand, in vitro results can be difficult to interpret because simplified systems introduce biases not present in cells. In this paper, the Ori-McKenny lab illustrates how powerful it is to combine in vitro and in vivo studies to investigate the mechanism of cellular processes.
MTs are essential to maintain the shape, polarity and intercellular transport in our neurons. The findings suggest that MAPs on the MTs coordinate with each other and that MT motors can interpret MAPs, which allows for the regulation of cargo transport. Monroy’s story only discussed MAP7 and tau, but MTs are also occupied by other MAPs. Also, MTs can be post-translationally regulated to favor or inhibit motor mobility. I am grateful that our neurons have a system to coordinate cargo transport, which makes up a powerful neural system that enables me to ponder the relationship between different MAPs, motors and how they are doing their jobs together on the MTs! I am excited to see what Monroy and Ori-McKenney come up with next! A more complex map about motors and MAPs is yet to be revealed!
Now, after sitting here for a while, I have decided to walk out to get a cup of iced tea. I suppose that the kinesin and dynein motors made their ways to the right destination and the long motor neuron axons delivered the message to my toes, since I made it to the long line and am enjoying it now.
References:
1. Burton PR (1988) Dendrites of mitral cell neurons contain microtubules of opposite polarity. Brain Res 473(1):107-115.
2. Ballatore C, Lee VM, & Trojanowski JQ (2007) Tau-mediated neurodegeneration in Alzheimer’s disease and related disorders. Nat Rev Neurosci 8(9):663-672.
3. Tymanskyj SR, Yang B, Falnikar A, Lepore AC, & Ma L (2017) MAP7 Regulates Axon Collateral Branch Development in Dorsal Root Ganglion Neurons. J Neurosci 37(6):1648-1661.
4. Ebneth A, et al. (1998) Overexpression of tau protein inhibits kinesin-dependent trafficking of vesicles, mitochondria, and endoplasmic reticulum: Implications for Alzheimer’s disease. Journal of Cell Biology 143(3):777-794.
5. Dixit R, Ross JL, Goldman YE, & Holzbaur EL (2008) Differential regulation of dynein and kinesin motor proteins by tau. Science 319(5866):1086-1089.
6. Barlan K, Lu W, & Gelfand VI (2013) The Microtubule-Binding Protein Ensconsin Is an Essential Cofactor of Kinesin-1. Current Biology 23(4):317-322.
7. Metzger T, et al. (2012) MAP and kinesin-dependent nuclear positioning is required for skeletal muscle function. Nature 484(7392):120-+.
8. Monroy BY, et al. (2018) Competition between microtubule-associated proteins directs motor transport. Nat Commun 9.
9. Ori-McKenney KM, Jan LY, & Jan YN (2012) Golgi Outposts Shape Dendrite Morphology by Functioning as Sites of Acentrosomal Microtubule Nucleation in Neurons. Neuron 76(5):921-930.






 Microtubule (MT) plus-end motor kinesin-1 and kinesin-3 are inhibited by tau patches, while tau has little effect on the MT minus-end motor dynein. MAP7 outcompetes tau and specifically enhances kinesin-1 movement. Similar to tau, MAP7 inhibits kinesin-3 but allows for the dynein movement.
Microtubule (MT) plus-end motor kinesin-1 and kinesin-3 are inhibited by tau patches, while tau has little effect on the MT minus-end motor dynein. MAP7 outcompetes tau and specifically enhances kinesin-1 movement. Similar to tau, MAP7 inhibits kinesin-3 but allows for the dynein movement.
 Comic about caseins coming together.
Comic about caseins coming together. A) Spores formed by roqueforti throughout blue cheese. B) Bloomy rind of brie formed by P. camemberti.
A) Spores formed by roqueforti throughout blue cheese. B) Bloomy rind of brie formed by P. camemberti.


 Image credit: “Piled Higher and Deeper” by Jorge Cham www.phdcomics.com
Image credit: “Piled Higher and Deeper” by Jorge Cham www.phdcomics.com Image credit: “Piled Higher and Deeper” by Jorge Cham www.phdcomics.com
Image credit: “Piled Higher and Deeper” by Jorge Cham www.phdcomics.com